According to new research from Rice University, widowed individuals experiencing intense grief after the loss of their spouse experience a significant increase in body inflammation following other stressful events.


According to new research from Rice University, widowed individuals experiencing intense grief after the loss of their spouse experience a significant increase in body inflammation following other stressful events.

Scholars, tourists, and locals eager to learn more about Rio de Janeiro have a new way to dive into its history with imagineRio, a digital platform that makes visible centuries of dramatic change in the city’s built environment. Supported by a grant from the Getty Foundation, the newly enhanced digital atlas will allow users to […]
Investments in science and technology research are vital to the United States’ economic growth and global leadership, according to a new report from Rice University’s Baker Institute for Public Policy. The Biden administration has made science and technology (S&T) a centerpiece of its early policy agenda with ambitious targets for federal investments in research and […]

Rice University bioengineers are using 3D printing and smart biomaterials to create an insulin-producing implant for Type 1 diabetics. The three-year project is a partnership between the laboratories of Omid Veiseh and Jordan Miller that’s supported by a grant from JDRF, the leading global funder of diabetes research. Veiseh and Miller will use insulin-producing […]
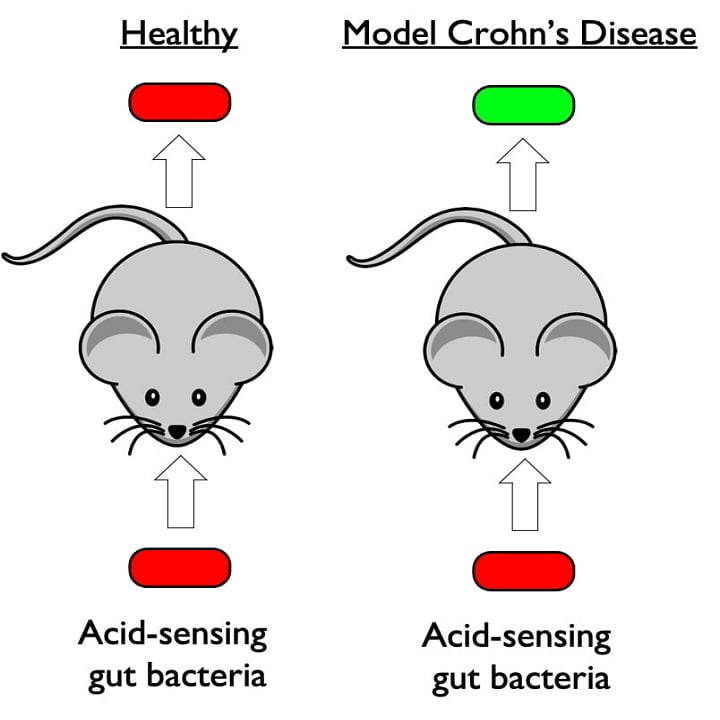
In an important step toward the clinical application of synthetic biology, Rice University researchers have engineered a bacterium with the necessary capabilities for diagnosing inflammatory bowel diseases. The engineered strain of the gut bacteria E. coli senses pH and glows when it encounters acidosis, an acidic condition that often occurs during flare ups of inflammatory […]
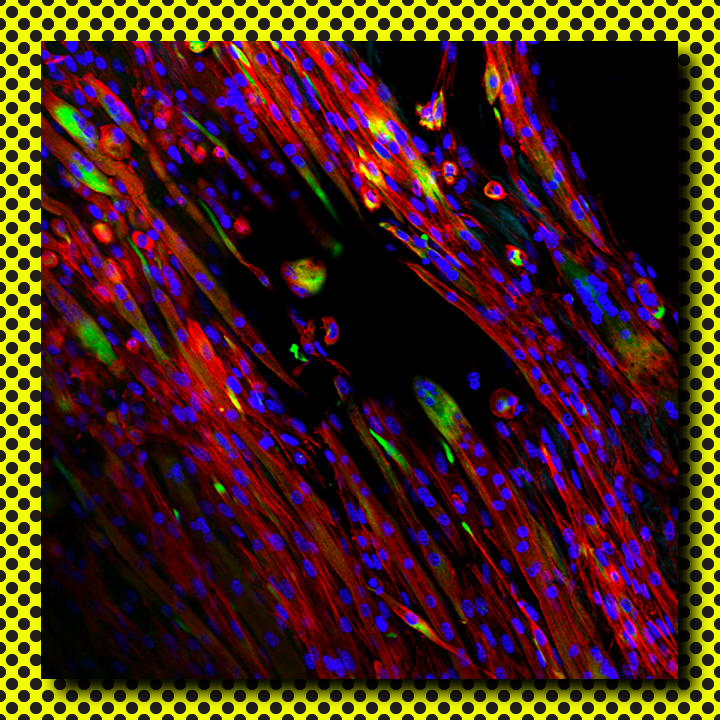
Bio-inspired scaffolds help promote muscle growth Rice University bioengineers adapt extracellular matrix for electrospinning Rice University bioengineers are fabricating and testing tunable electrospun scaffolds completely derived from decellularized skeletal muscle to promote the regeneration of injured skeletal muscle. Their paper in Science Advances shows how natural extracellular matrix can be made to mimic native skeletal muscle and direct the […]

Wearable glucose monitors shed light on progression of Type 2 diabetes in Hispanic/Latino adults Study by Sansum Diabetes Research Institute and Rice University points to new directions for improved diabetes care In one of the first studies of its kind, medical and engineering researchers have shown wearable devices that continuously monitor blood sugar provide new […]
Houston Methodist, Rice U. launch neuroprosthetic collaboration Center for Translational Neural Prosthetics and Interfaces to focus on restoring brain function after disease, injury Neurosurgery’s history of cutting diseases out of the brain is morphing into a future in which implanting technology intothe brain may help restore function, movement, cognition and memory after patients suffer strokes, […]

TB-causing bacteria remember prior stress, react quickly to new stress Tuberculosis bacteria have evolved to remember stressful encounters and react quickly to future stress, according to a study by computational bioengineers at Rice University and infectious disease experts at Rutgers New Jersey Medical School (NJMS). Published online in the open-access journal mSystems, the research identifies a […]
Michael Stern and James McNew (Photo by Jeff Fitlow/Rice University) Study: Early, late stages of degenerative diseases are distinct Two-phase theory applies to diseases like Alzheimer’s, Parkinson’s, muscle atrophy Rice University biochemists Michael Stern and James McNew have studied how neurodegeneration kills cells. They’ve conducted countless experiments over more than a decade, and they’ve summarized […]